Team 11 : reMoVE-B
Molecular Vulnerabilities of Tumor Escape in mature B-cell Malignancies
Team leaders : Catherine Pellat and David Chiron
Research projects
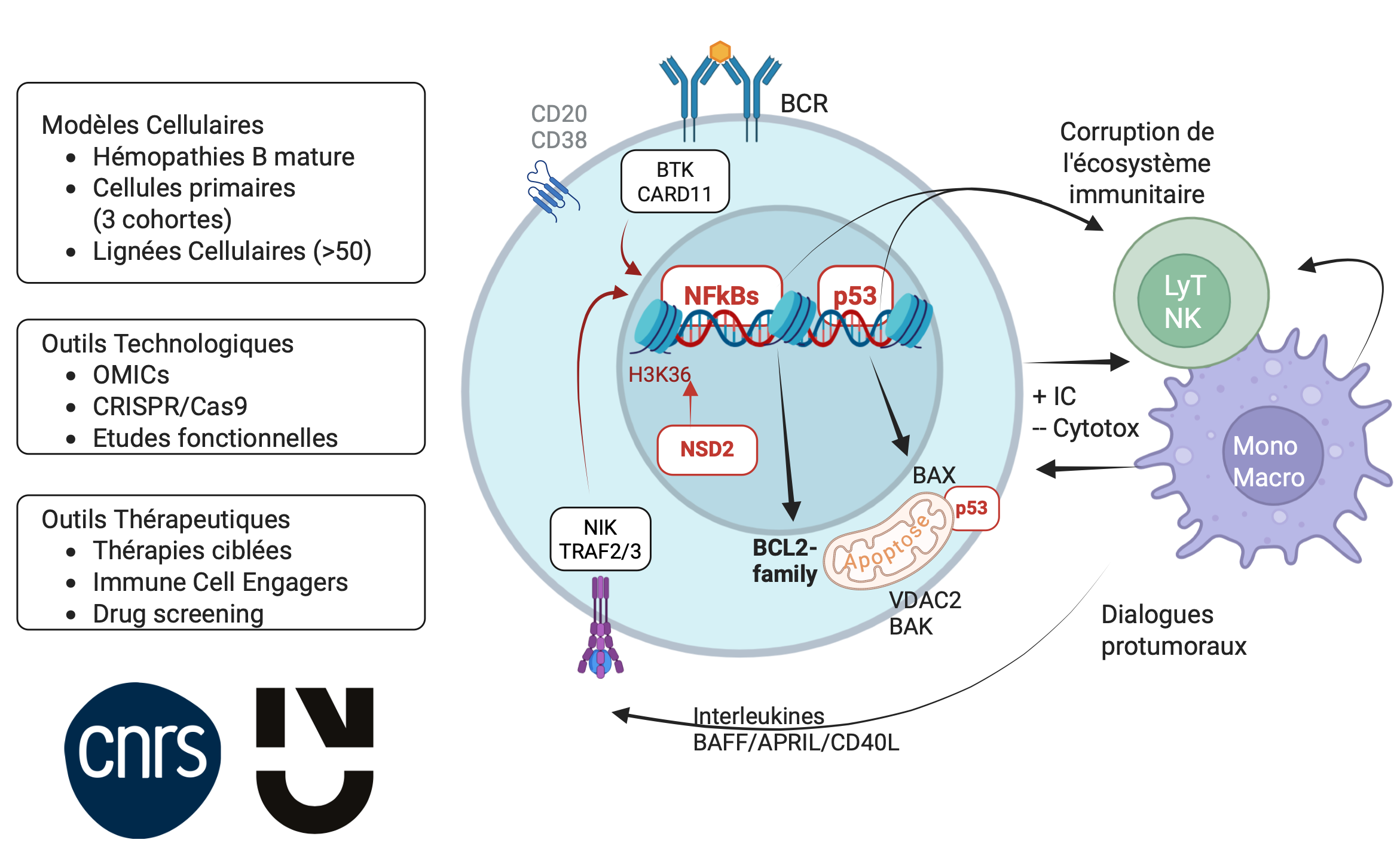
Hallmarks of cancer, which include dysregulation of cell cycle and apoptosis are both of tumor and of microenvironment origins. Although these hallmarks favor cancer survival and growth, they also confer specific addictions or vulnerabilities to cancer cells that can be exploited for therapies with regard to cancer heterogeneity and specificity.
We work on identifying vulnerabilities – Achille’s heel – that are consequent to the adaptation of cancer cells to survival and resistance within their ecosystem. More precisely, our project builds upon our previous work aimed at elucidating the molecular mechanisms underlying therapeutic resistance within tumor ecosystems (see Main publications).
Now, we aim to explore novel aspects of resistance pathways, structured around three complementary research axes:
Transcriptional Alterations and Epigenetic Deregulation: We will investigate transcriptional changes driven by epigenetic mechanisms that disrupt these pathways.
Signaling Perturbations via Mutant Proteins: We will study alterations in intracellular signaling by characterizing the abnormal functions of mutant proteins (interactome, subcellular localization).
Functional Impact on Apoptosis and Immune Evasion: We aim to elucidate the functional consequences of pathway dysregulation on apoptosis resistance (BCL2 family) and the disruption of the immune microenvironment.
This integrative approach is designed to identify novel therapeutic targets in refractory B-cell malignancies.
High throughput technologies to uncover novel Achille’s heel
In addtion to supervised analyses based on gold standard biochemestry, cellular and molecular biology, our teams have acces to high throughput technologies such as single cell RNA-seq and Cut n Run. All the skills needed to analyze the big data generated are also mastered within the team.
Functional validation based on cell Models and Patient Cohorts
In collaboration with the Hematology Department of Nantes, we have established two large real-world patient cohorts with multiple myeloma (MM) and mantle cell lymphoma (MCL) (over 600 patients), with full clinico-biological annotation. From these patient samples, we have generated robust primary cells (co)culture models as well as two molecularly and functionally characterized cell line panels, from which we derive isogenic models—either knock-out (KO) for loss-of-function analysis or knock-in (KI) for mutant expression.
Contact
Centre de Recherche en Cancérologie et Immunologie Intégrée Nantes Angers
⌂ Institut de Recherche en Santé de l'Université de Nantes - 8 quai Moncousu - BP 70721 - 44007 Nantes cedex 1
✉ catherine.pellat-deceunynck@univ-nantes.fr
✉ david.chiron@univ-nantes.fr
✆ +332 28 08 02 98


Team certified on 02 august 2024
Management of biological resources for Multiple Myeloma and Mantle Cell Lymphoma :
1- Processing and qualification of patient sample cells and associated data (excluding clinical data)
2- Processing and qualification of cell lines and associated data
Members
|
Catherine Pellat, DR CNRS
David Chiron, DR CNRS Agnès Moreau-Aubry, PU Antonin Papin, MCU Cyrille Touzeau, PU-PH Marion Eveillard, PU-PH Benoît Tessoulin, PU-PH Yannick Le Bris, PH Anne Lok, PH Jessie Bourcier, PH Nicoletta Libera Lilli, Cheffe de projet translationnel Yanis Macé, Post-doctorant Géraldine Descamps, IE Christelle Dousset, IE Sophie Maiga, AI Philippine Giowachini, AI Céline Bellanger, IE Louise Hervouet, Doctorante Candice Madiot, Doctorante Camille Glemarec, Doctorant Ivan Anguilet, Doctorant Elyne Roux, M2 Solène Sepré, M2 Martin Bauer, M2 |
 |
Durand R, Descamps G, Bellanger C, Dousset C, Maïga S, Alberge JB, Derrien J, Cruard J, Minvielle S, Lilli NL, Godon C, Le Bris Y, Tessoulin B, Amiot M, Gomez-Bougie P, Touzeau C, Moreau P, Chiron D, Moreau-Aubry A, Pellat-Deceunynck C. A p53 score derived from TP53 CRISPR/Cas9 HMCLs predicts survival and reveals a major role of BAX in the response to BH3 mimetics.Blood. (2024)
Sarkozy C, Tessoulin B, Chiron D. Unraveling MCL biology to understand resistance and identify vulnerabilities. Blood. (2024)
Durand R, Bellanger C, Kervoëlen C, Tessoulin B, Dousset C, Menoret E, Asnagli H, Parker A, Beer P, Pellat-Deceunynck C, Chiron D. Selective pharmacologic targeting of CTPS1 shows single-agent activity and synergizes with BCL2 inhibition in aggressive mantle cell lymphoma. Haematologica. (2024)
Decombis S, Bellanger C, Le Bris Y, Madiot C, Jardine J, Santos JC, Boulet D, Dousset C, Menard A, Kervoelen C, Douillard E, Moreau P, Minvielle S, Moreau-Aubry A, Tessoulin B, Roue G, Bidère N, Le Gouill S, Pellat-Deceunynck C, Chiron D. CARD11 gain of function upregulates BCL2A1 expression and promotes resistance to targeted therapies combination in B-cell lymphoma. Blood. (2023)
Decombis S, Papin A, Bellanger C, Sortais C, Dousset C, Le Bris Y, Riveron T, Blandin S, Hulin P, Tessoulin B, Rouel M, Le Gouill S, Moreau-Aubry A, Pellat-Deceunynck C, Chiron D. The IL32/BAFF axis supports prosurvival dialogs in the lymphoma ecosystem and is disrupted by NIK inhibition. Haematologica. (2022)
Le Gouill S, Morschhauser F, Chiron D, Bouabdallah K, Cartron G, Casasnovas O, Bodet-Milin C, Ragot S, Bossard C, Nadal N, Herbaux C, Tessoulin B, et al. Ibrutinib, obinutuzumab, and venetoclax in relapsed and untreated patients with mantle cell lymphoma: a phase 1/2 trial. Blood. (2021)
Seiller C, Maiga S, Touzeau C, Bellanger C, Kervoëlen C, Descamps G, Maillet L, Moreau P, Pellat-Deceunynck C, Gomez-Bougie P, Amiot M. Dual targeting of BCL2 and MCL1 rescues myeloma cells resistant to BCL2 and MCL1 inhibitors associated with the formation of BAX/BAK hetero-complexes. Cell Death Dis. (2020)
Papin A, Tessoulin B, Bellanger C, Moreau A, Le Bris Y, Maisonneuve H, Moreau P, Touzeau C, Amiot M, Pellat-Deceunynck C, Le Gouill S, Chiron D. CSF1R and BTK inhibitions as novel strategies to disrupt the dialog between mantle cell lymphoma and macrophages. Leukemia. (2019)
Lok A, Descamps G, Tessoulin B, Chiron D, Eveillard M, Godon C, Le Bris Y, Vabret A, Bellanger C, Maillet L, Barillé-Nion S, Gregoire M, Fonteneau JF, Le Gouill S, Moreau P, Tangy F, Amiot M, Moreau-Aubry A, Pellat-Deceunynck C. p53 regulates CD46 expression and measles virus infection in myeloma cells. Blood Adv. (2018)
Gomez-Bougie P, Maiga S, Tessoulin B, Bourcier J, Bonnet A, Rodriguez MS, Le Gouill S, Touzeau C, Moreau P, Pellat-Deceunynck C, Amiot M. BH3-mimetic toolkit guides the respective use of BCL2 and MCL1 BH3-mimetics in myeloma treatment. Blood. (2018)
Le Gouill S, Thieblemont C, Oberic L, et al., Rituximab after Autologous Stem-Cell Transplantation in Mantle-Cell Lymphoma. NEJM. (2017)
Leverson JD, Sampath D, Souers AJ, Rosenberg SH, Fairbrother WJ, Amiot M et al., Found in Translation: How Preclinical Research Is Guiding the Clinical Development of the BCL2-Selective Inhibitor Venetoclax. Cancer Discov. (2017)
Touzeau C, Le Gouill S, Mahé B, Boudreault JS, Gastinne T, Blin N, Caillon H, Dousset C, Amiot M, Moreau P. Deep and sustained response after venetoclax therapy in a patient with very advanced refractory myeloma with translocation t(11;14). Haematologica. (2017)
Chiron D, Bellanger C, Papin A, Tessoulin B, Dousset C, Maïga S, Moreau A, Esbelin J, Trichet V, Chen-Kiang S, Moreau P, Touzeau C, Le Gouill S, Amiot M, Pellat-Deceunynck C. Rational targeted therapies to overcome microenvironment-dependent expansion of mantle cell lymphoma. Blood. (2016)
Touzeau C, Ryan J, Guerriero J, Moreau P, Ni Chonghaile T, Le Gouill S, Richardson P, Anderson K, Amiot M, Letai A. BH3-profiling identifies heterogeneous dependency on Bcl-2 family members in Multiple Myeloma and predicts sensitivity to BH3 mimetics. Leukemia. (2015)
Chiron D, Di Liberto M, Martin P, Huang X, Sharman J, Blecua P, Mathew S, Vijay P, Eng K, Ali S, Johnson A, Chang B, Ely S, Elemento O, Mason CE, Leonard JP, Chen-Kiang S. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov. (2014)
Tessoulin B, Descamps G, Moreau P, Maiga S, Lodé L, Godon C, Marionneau-Lambot S, Ouiller T, Le Gouill S, Amiot M, Pellat-Deceunynck C. PRIMA-1Met induces myeloma cell death independently of p53 by impairing the redox balance. Blood. (2014)
Touzeau C, Dousset C, Le Gouill S, Sampath D, Leverson JD, Souers AJ, Maïga S, Béné MC, Moreau P, Pellat-Deceunynck C, Amiot M. The Bcl-2 specific BH3 mimetic ABT-199 : a promising targeted therapy for t(11 ;14) multiple myeloma. Leukemia. (2014)




